Overview

Bacteria are everywhere, and their interactions with each other, their hosts, and the environment can be both beneficial and harmful. While pathogens cause disease and antibiotic resistance makes infections harder to treat, probiotics offer opportunities for new therapies and environmental solutions. Advances in synthetic biology, genomics, and protein engineering now allow us to make and harness fundamental insights into bacteria–bacteria, bacteria–host, and bacteria–environment interactions. With these tools, we can design microbes and their products to prevent infection, treat disease, and promote sustainability. Our work spans all these areas, unified by their impact on human health and the future of our planet. Current projects include:

Uncovering microcin diversity and functions
Microcins are potent, selective antibacterial peptides that play key roles in microbial competition. For decades, little was known about these intriguing molecules. We are now developing computational and experimental approaches to uncover and characterize the hidden diversity of microcins across Gram-negative bacteria. Our work investigates their unique functions, rare ability to translocate across bacterial membranes, interactions with hosts, and potential to target pathogens and shape microbiomes.
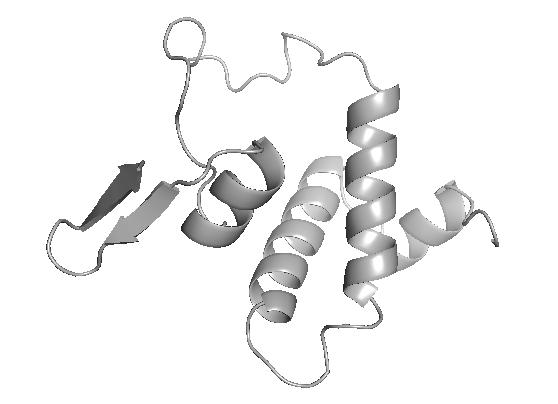
Designing bacteria-host interactions
Bacteria colonize diverse host niches and interact with resident microbes across human, animal, and environmental microbiomes. We aim to harness these natural interactions and engineer beneficial bacteria to deliver therapeutic proteins and genes. Such designs can enable infection treatment, microbiome modulation, immune system control, and enhanced environmental resilience.

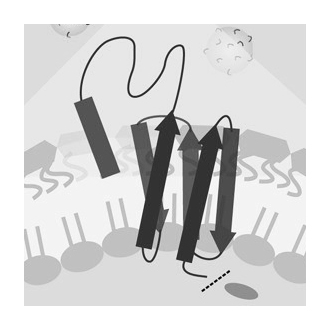
Building bacterial display and secretion systems
Bacteria interact with their environment via proteins that are secreted to the extracellular space or presented on the cell surface. We advance systems for cellular protein secretion and display to discover, engineer, and deliver both natural and synthetic bioactive proteins. These platforms enable high-throughput analysis of otherwise inaccessible proteins and generate datasets that accelerate computational approaches to protein discovery and design.
Developing peptide and nanobody effectors
Peptides and antibodies serve as versatile chemical scaffolds to target a wide range of biological molecules. We develop molecular screening and computational strategies to accelerate discovery of functional variants and to elucidate how sequence influences biological activity. These protein candidates open new pathways for treating infection, modulating host immunity, and advancing environmental remediation.
